Paranoid? Masking, anonymizing, and obfuscating PII in XML and JSON data
Data masking, anonymization, and obfuscation are methods to scramble personally identifiable information (PII). We can apply different techniques to mask data, e.g. you may substitute values (swap na for another) or randomly substitute characters just preserving the data type.
There are no open source tools (at least we are not aware of any) that automate masking of data in XML or JSON. That has changed as of today. We are releasing Paranoid an open source tool to mask XML and JSON data.
Pypi link – https://pypi.org/project/PARANOID/
Bitbucket – https://bitbucket.org/sonra/paranoid/src/master/
Use Flexter to turn XML and JSON into Valuable Insights
- 100% Automation
- 0% Coding
Paranoid and Flexter
Paranoid is best used in combination with Flexter.
Flexter, is an enterprise XML conversion tool. It converts XML and JSON to text, a relational database or Hadoop/Spark (ORC, Parquet, Avro). If you have privacy concerns around uploading your data you can now download and use Paranoid to mask individual elements.
Paranoid is developed in Python. You will need Python 2.6+ to use it. You can either mask all of the elements in an individual XML/JSON, a folder of XML/JSON, or just individual elements. Paranoid replaces sensitive data with random characters. It can’t be decoded.
Here is a list of the Paranoid switches:
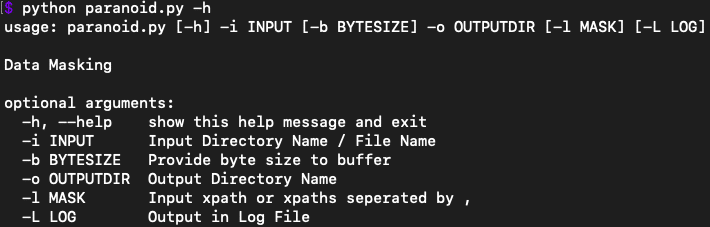
If you are still concerned with privacy you can always check the script yourself (Remember it is an open source tool after all).
Paranoid masking in action
In this blog we will show you how Paranoid and Flexter work hand in hand to mask and convert data in XML.
Anonymising all elements in a single XML document
In a first step we will mask an individual XML element. Let’s look at a sample XML file before masking.
Let’s use Paranoid in a next step to mask all of the XML elements in this sample document.

We can see the argument -I given for our input file sample.xml and argument -o for the output location.
Now let’s look at the head of the xml file before and after masking. All of the XML elements (apart from namespaces) have been obfuscated with random characters.
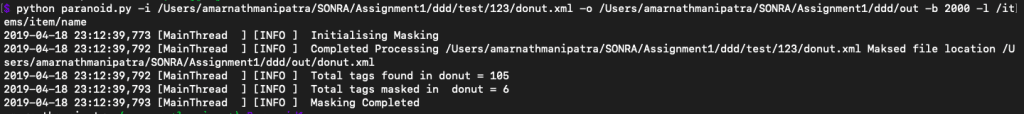
We can also have a look at stats such as number of xml tags found and number of tags that have been masked
Original File(SAMPLE)(Complete files you will find below)
|
1 2 3 4 5 6 7 8 |
<clinical_study> <!-- This xml conforms to an XML Schema at: https://clinicaltrials.gov/ct2/html/images/info/public.xsd --> <required_header> <download_date>ClinicalTrials.gov processed this data on March 28, 2019</download_date> <link_text>Link to the current ClinicalTrials.gov record.</link_text> <url>https://clinicaltrials.gov/show/NCT00000102</url> </required_header> |
Mask File(SAMPLE)
|
1 2 3 4 5 6 7 8 |
<clinical_study> <!-- This xml conforms to an XML Schema at: https://clinicaltrials.gov/ct2/html/images/info/public.xsd --> <required_header> <download_date>EakznbqmWsqeja.lrp wddsydnef vxfo erdh fa Qbmcx 43, 7483</download_date> <link_text>Idbi ee wum zvsbemg ZvhsgiviRjyfyp.ehd imiskk.</link_text> <url>ytntg://qorczrjhnayhcp.ekm/nndw/GUZ23553334</url> </required_header> |
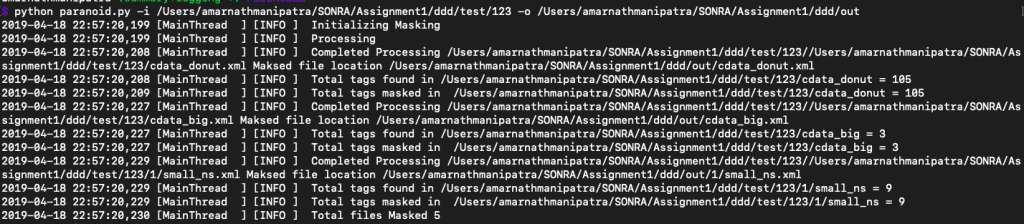
Masking all elements in multiple JSON/XML documents
Well all this looks pretty cool, but what if you want all the files in a directory/sub directory to be masked? What if the directory contains both JSON and XML?
Not to worry. Paranoid can also handle this requirement.
Apart from the stats you obtain an additional stats of number of files processed in the process.
|
1 |
paranoid -i /Users/amarnathmanipatra/SONRA/ddd/test/AllPublicXML/NCT0000xxxx/ -o /Users/amarnathmanipatra/SONRA/ddd/test/out |
Masking large XML files
This is all great. But what if you have a large file and less memory?
Well Paranoid has a solution for this as well. It offers a switch -b where you can provide a byte size and Paranoid will read the specified size mentioned by you at a moment and mask those contents at the point. Let’s demonstrate it.
|
1 |
paranoid -i /Users/amarnathmanipatra/SONRA/ddd/test/AllPublicXML/NCT0000xxxx/NCT00000102.xml -o /Users/amarnathmanipatra/SONRA/ddd/test/out -b 2000 |
That’s right paranoid just buffered 2000 bytes and streamed the data to the output location.
Masking individual tags in an XML document
There is one more thing we wanted to show you. You can be selective in the tags you mask.
You can provide a comma separated list of XPaths to mask.
|
1 |
paranoid -i /Users/amarnathmanipatra/SONRA/ddd/test/AllPublicXML/NCT0000xxxx/NCT00000102.xml -o /Users/amarnathmanipatra/SONRA/ddd/test/out -b 2000 -l /clinical_study/required_header/download_date,/clinical_study/required_header/url |
We are masking two tags: download_date and url
Original File
|
1 2 3 4 5 6 7 8 |
<clinical_study> <!-- This xml conforms to an XML Schema at: https://clinicaltrials.gov/ct2/html/images/info/public.xsd --> <required_header> <download_date>ClinicalTrials.gov processed this data on March 28, 2019</download_date> <link_text>Link to the current ClinicalTrials.gov record.</link_text> <url>https://clinicaltrials.gov/show/NCT00000102</url> </required_header> |
Mask File
|
1 2 3 4 5 6 7 8 |
<clinical_study> <!-- This xml conforms to an XML Schema at: https://clinicaltrials.gov/ct2/html/images/info/public.xsd --> <required_header> <download_date>QbjytvupUowrfh.hpt bojjhgjyw prdb zfcq bv Pfpzt 17, 5147</download_date> <link_text>Link to the current ClinicalTrials.gov record.</link_text> <url>btuqg://cbxhegfnhecfbx.nax/qndh/KNZ39335989</url> </required_header> |
Storing the output in Log File
Wondering how you can store the output to reuse it later. We are providing a log function where with the help of -L argument you can store the output of it in a log file.

Sample image of the log file
Using Flexter on masked and original data
For data analytics it is typically better to have data in a relational form. Querying data in XML is hard or near impossible. SQL is the lingua franca for data. Flexter is the perfect tool to convert your masked data to a relational format. The premium version also supports conversion of files to a database or popular Hadoop formats, e.g. ORC, Parquet, Avro. You can find out more about Flexter in our product and FAQ section. Please refer to our blog post Converting clinical trials XML data to Snowflake and Quicksight with Flexter.
Let’s have a look at the output of Flexter before masking
Here is the output of Flexter after masking. Random characters
If you’re looking to load JSON data into a database for analysis, Flexter provides an automated workflow that transforms complex JSON into structured tables, ready for integration with your target system. Learn more in our Flexter JSON to SQL Guide